728x90
반응형
SMALL
from sklearn.datasets import make_classification, load_breast_cancer
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import OneHotEncoder, LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.compose import make_column_transformer
import matplotlib
matplotlib.rcParams["font.family"]="Malgun Gothic"
matplotlib.rcParams["axes.unicode_minus"]= False
x_data = np.array( [[25.8],[26.6],[28.1],[29.0],[30.5],
[31.0],[33.6],[39.3],[43.3],[45.8] ] )
y_data = np.array( [[1],[0],[0],[1],[1],[1],[1],[0],[0],[1] ] )
df = pd.DataFrame( x_data, columns=['BMI'] )
df['당뇨여부'] = y_data
df

confusion matrix
y_pred = [1,1,1,1,1,1,1,1,1,1]
cm = metrics.confusion_matrix(y_data,y_pred)
cm[OUT]:
array([[0, 4],
[0, 6]], dtype=int64)
y_pred = [0,1,1,1,1,1,1,1,1,1]
cm = metrics.confusion_matrix(y_data,y_pred)
cm[OUT]:
array([[0, 4],
[1, 5]], dtype=int64)
# y_data = [1,0,0,1,1,1,1,0,0,1]
y_pred = [0,1,1,1,1,1,1,1,1,1]
cm = metrics.confusion_matrix(y_data,y_pred)
sns.heatmap(cm,annot=True,cmap='Reds',xticklabels=['정상판정','당뇨판정'],yticklabels=['정상','당뇨환자'] )
plt.show()
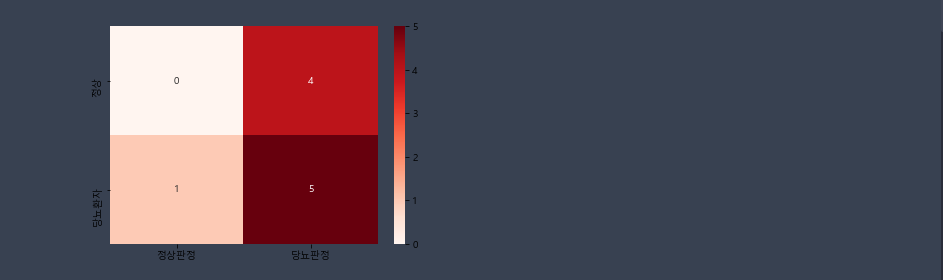
# y_data = [1,0,0,1,1,1,1,0,0,1]
y_pred = [0,0,1,1,1,1,1,1,1,1]
cm = metrics.confusion_matrix(y_data,y_pred)
sns.heatmap(cm,annot=True,cmap='Reds',xticklabels=['정상판정','당뇨판정'],yticklabels=['정상','당뇨환자'] )
plt.show() # TN FP FN TP (Z방향)

정밀도(Precision)
- 모델이 True로 예측한 데이터 중 실제로 True인 데이터 비율
- TP/(TP+FP)
# 1
precision = 5/(5+3)
precision[OUT]:
0.625
# 2
metrics.precision_score(y_data,y_pred)[OUT]:
0.625재현율(Recall)
- 실제로 True인 데이터 중 모델이 True로 예측한 데이터 비율
- TP/(TP+FN)
# 1
recall = 5/(5+1)
recall[OUT]:
0.8333333333333334
# 2
metrics.recall_score(y_data,y_pred)[OUT]:
0.8333333333333334F1 score
- 정밀도(Precision), 재현율(Recall) 의 조화평균
- 2/(1/Precision + 1/Recall)
# 1-1
f1score = 2/(1/precision+1/recall)
f1score[OUT]:
0.7142857142857143
# 1-2
f1score = 2/((5+1)/5+(5+3)/5)
f1score[OUT]:
0.7142857142857143
# 2
metrics.f1_score(y_data,y_pred)[OUT]:
0.7142857142857143데이터 불러오기 (pd.read_csv)
python 파일 경로에 data5 폴더 만든 후 다음의 'pima-indians-diabetes.data.csv'파일 넣어놓기
pima-indians-diabetes.data.csv
0.02MB
연습문제
피마인디안 데이터를 이용하여 confusion matrix(heatmap),precision,recall,f1 score를 구하시오
Solution
df = pd.read_csv('data5/pima-indians-diabetes.data.csv')
df.head(3)

# 피쳐와 레이블 지정 및 학습, 테스트 데이터셋 설정
x_data = df.iloc[:,:-1]
y_data = df.iloc[:,-1]
x_train, x_test, y_train, y_test = train_test_split(x_data, y_data, test_size=0.2,random_state=1,
stratify=y_data) # stratify=y_data 반드시 (0,1균형)
# gridsearchcv로 최적의 파라메터값 구하기
model_logistic = make_pipeline( StandardScaler(), LogisticRegression() )
param_value = {'logisticregression__C': np.linspace(0,50, 100),'logisticregression__penalty':['l1', 'l2', 'elasticnet', 'none']
, 'logisticregression__solver': ['newton-cg', 'lbfgs', 'liblinear', 'sag', 'saga']}
model_logistic = GridSearchCV(model_logistic, param_grid = param_value,scoring='f1')
model_logistic.fit(x_train, y_train)[OUT]:
GridSearchCV(cv=None, error_score=nan,
estimator=Pipeline(memory=None,
steps=[('standardscaler',
StandardScaler(copy=True,
with_mean=True,
with_std=True)),
('logisticregression',
LogisticRegression(C=1.0,
class_weight=None,
dual=False,
fit_intercept=True,
intercept_scaling=1,
l1_ratio=None,
max_iter=100,
multi_class='auto',
n_jobs=None,
penalty='l2',
random_state=No...
42.92929293, 43.43434343, 43.93939394, 44.44444444, 44.94949495,
45.45454545, 45.95959596, 46.46464646, 46.96969697, 47.47474747,
47.97979798, 48.48484848, 48.98989899, 49.49494949, 50. ]),
'logisticregression__penalty': ['l1', 'l2',
'elasticnet', 'none'],
'logisticregression__solver': ['newton-cg', 'lbfgs',
'liblinear', 'sag',
'saga']},
pre_dispatch='2*n_jobs', refit=True, return_train_score=False,
scoring='f1', verbose=0)
print(model_logistic.best_params_)
print(model_logistic.best_score_)
print(model_logistic.best_estimator_)[OUT]:
{'logisticregression__C': 0.5050505050505051, 'logisticregression__penalty': 'l2', 'logisticregression__solver': 'liblinear'}
0.6427440123049879
Pipeline(memory=None,
steps=[('standardscaler',
StandardScaler(copy=True, with_mean=True, with_std=True)),
('logisticregression',
LogisticRegression(C=0.5050505050505051, class_weight=None,
dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None,
max_iter=100, multi_class='auto',
n_jobs=None, penalty='l2',
random_state=None, solver='liblinear',
tol=0.0001, verbose=0, warm_start=False))],
verbose=False)
# predict
y_pred_train= model_logistic.predict(x_train)
y_pred_test = model_logistic.predict(x_test)
# train confusion_matrix
cm2_train = metrics.confusion_matrix( y_train, y_pred_train )
sns.heatmap( cm2_train, annot=True, cmap='Reds',
xticklabels=['정상판정','당뇨판정'],yticklabels=['정상','당뇨환자'] )
plt.show()

# test confusion_matrix
cm_test = metrics.confusion_matrix( y_test, y_pred_test )
sns.heatmap( cm_test, annot=True, cmap='Reds',
xticklabels=['정상판정','당뇨판정'],yticklabels=['정상','당뇨환자'] )
plt.show()

# precision, recall, f1 score
print('train 정밀도: ',metrics.precision_score( y_train, y_pred_train ))
print('train 재현율: ',metrics.recall_score( y_train, y_pred_train ))
print('train F1 score: ',metrics.f1_score( y_train, y_pred_train ))
print('test 정밀도: ',metrics.precision_score( y_test, y_pred_test ))
print('test 재현율: ',metrics.recall_score( y_test, y_pred_test ))
print('test F1 score: ',metrics.f1_score( y_test, y_pred_test ))[OUT]:
train 정밀도: 0.7398843930635838
train 재현율: 0.5981308411214953
train F1 score: 0.6614987080103358
test 정밀도: 0.7631578947368421
test 재현율: 0.5370370370370371
test F1 score: 0.6304347826086957False Positive Rate(FPR)와 True Positive Rate(TPR)
- True Positive는 실제로 이 사람은 암에 걸려있고, 암에 걸렸다고 판단한 것
- False Positive는 실제로는 암에 걸리지 않았는데도 암에 걸렸다고 잘못 판단한 것
proba = model_logistic.predict_proba(x_test)
proba[OUT]:
array([[0.92149458, 0.07850542],
[0.82769673, 0.17230327],
[0.95368836, 0.04631164],
[0.54904432, 0.45095568],
[0.7037808 , 0.2962192 ],
[0.76450919, 0.23549081],
[0.9666442 , 0.0333558 ],
[0.89781377, 0.10218623],
[0.22487114, 0.77512886],
[0.9477928 , 0.0522072 ],
[0.77896917, 0.22103083],
[0.94182684, 0.05817316],
[0.25086861, 0.74913139],
[0.9673507 , 0.0326493 ],
[0.9469779 , 0.0530221 ],
[0.70492167, 0.29507833],
[0.72160399, 0.27839601],
[0.48129855, 0.51870145],
[0.84703038, 0.15296962],
[0.30864599, 0.69135401],
[0.25013187, 0.74986813],
[0.90954642, 0.09045358],
[0.56188213, 0.43811787],
[0.96666285, 0.03333715],
[0.76090456, 0.23909544],
[0.81698123, 0.18301877],
[0.53051664, 0.46948336],
[0.90889639, 0.09110361],
[0.94806603, 0.05193397],
[0.92665124, 0.07334876],
[0.93358053, 0.06641947],
[0.99610479, 0.00389521],
[0.96920719, 0.03079281],
[0.76091205, 0.23908795],
[0.3055346 , 0.6944654 ],
[0.13950067, 0.86049933],
[0.82270774, 0.17729226],
[0.14230345, 0.85769655],
[0.81077442, 0.18922558],
[0.84587552, 0.15412448],
[0.90404994, 0.09595006],
[0.13854116, 0.86145884],
[0.85271117, 0.14728883],
[0.53797422, 0.46202578],
[0.75095504, 0.24904496],
[0.76673392, 0.23326608],
[0.75634739, 0.24365261],
[0.87339868, 0.12660132],
[0.48362853, 0.51637147],
[0.08125913, 0.91874087],
[0.95713645, 0.04286355],
[0.80237839, 0.19762161],
[0.34902836, 0.65097164],
[0.59715003, 0.40284997],
[0.77098621, 0.22901379],
[0.69846428, 0.30153572],
[0.83557302, 0.16442698],
[0.96391927, 0.03608073],
[0.34530612, 0.65469388],
[0.3124526 , 0.6875474 ],
[0.93532389, 0.06467611],
[0.74862647, 0.25137353],
[0.96673382, 0.03326618],
[0.90674122, 0.09325878],
[0.02022432, 0.97977568],
[0.89268208, 0.10731792],
[0.99784427, 0.00215573],
[0.19363839, 0.80636161],
[0.96056442, 0.03943558],
[0.59723521, 0.40276479],
[0.38505908, 0.61494092],
[0.95321852, 0.04678148],
[0.61123046, 0.38876954],
[0.58431949, 0.41568051],
[0.49844719, 0.50155281],
[0.78285071, 0.21714929],
[0.26944796, 0.73055204],
[0.5282177 , 0.4717823 ],
[0.67818216, 0.32181784],
[0.51060168, 0.48939832],
[0.66289239, 0.33710761],
[0.55367312, 0.44632688],
[0.53212045, 0.46787955],
[0.52438343, 0.47561657],
[0.65570145, 0.34429855],
[0.38943805, 0.61056195],
[0.76897902, 0.23102098],
[0.65440993, 0.34559007],
[0.12026494, 0.87973506],
[0.74665126, 0.25334874],
[0.29931528, 0.70068472],
[0.886628 , 0.113372 ],
[0.75201217, 0.24798783],
[0.63371868, 0.36628132],
[0.78760029, 0.21239971],
[0.76562326, 0.23437674],
[0.89259515, 0.10740485],
[0.98343699, 0.01656301],
[0.34312938, 0.65687062],
[0.59355311, 0.40644689],
[0.75423653, 0.24576347],
[0.92583614, 0.07416386],
[0.60383329, 0.39616671],
[0.06268092, 0.93731908],
[0.68770458, 0.31229542],
[0.558347 , 0.441653 ],
[0.32181097, 0.67818903],
[0.88412902, 0.11587098],
[0.88319545, 0.11680455],
[0.04325884, 0.95674116],
[0.60578982, 0.39421018],
[0.90556764, 0.09443236],
[0.89032483, 0.10967517],
[0.5990761 , 0.4009239 ],
[0.29057605, 0.70942395],
[0.15814727, 0.84185273],
[0.58953068, 0.41046932],
[0.89759334, 0.10240666],
[0.0926567 , 0.9073433 ],
[0.24594301, 0.75405699],
[0.73149413, 0.26850587],
[0.99016527, 0.00983473],
[0.91650503, 0.08349497],
[0.71284315, 0.28715685],
[0.83767829, 0.16232171],
[0.34981299, 0.65018701],
[0.88398825, 0.11601175],
[0.92629751, 0.07370249],
[0.44491645, 0.55508355],
[0.35669516, 0.64330484],
[0.25089254, 0.74910746],
[0.87851809, 0.12148191],
[0.92566285, 0.07433715],
[0.79985149, 0.20014851],
[0.92961655, 0.07038345],
[0.73456801, 0.26543199],
[0.38378094, 0.61621906],
[0.79912859, 0.20087141],
[0.89452395, 0.10547605],
[0.40957796, 0.59042204],
[0.96941315, 0.03058685],
[0.8512037 , 0.1487963 ],
[0.86981501, 0.13018499],
[0.9108733 , 0.0891267 ],
[0.66538102, 0.33461898],
[0.83312963, 0.16687037],
[0.94867698, 0.05132302],
[0.63925238, 0.36074762],
[0.60618884, 0.39381116],
[0.81419883, 0.18580117],
[0.25664574, 0.74335426],
[0.8989794 , 0.1010206 ],
[0.45106973, 0.54893027],
[0.94524806, 0.05475194]])
rr = pd.DataFrame([y_test])
rr = rr.T
rr['proba'] = proba[:,1]
rr

fpr,tpr,threasholds = metrics.roc_curve(y_test,proba[:,1])
plt.plot(fpr,tpr,'r--',label='logistic')
plt.plot([0,1],[0,1],'k--',label='prediction')
plt.xlabel('FPR')
plt.ylabel('TPR')
plt.legend()
plt.show()
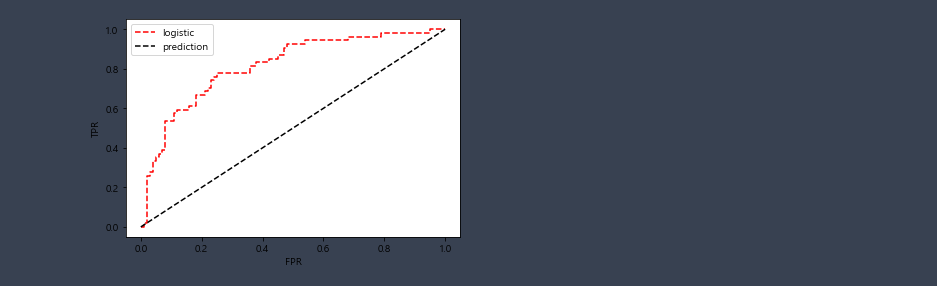
review
- confusion matrix, precision, recall, f1 score, ROC curve
728x90
반응형
LIST
'코딩으로 익히는 Python > 모델링' 카테고리의 다른 글
| [Python] 12. 다중분류 (0) | 2021.01.21 |
|---|---|
| [Python] 11. softmax (4) | 2021.01.21 |
| [Python] 9. Logistic Regression (로지스틱 회귀) (0) | 2021.01.20 |
| [Python] 8. Sigmoid & Logistic (0) | 2021.01.20 |
| [Python] 7. Sigmoid 함수 (0) | 2021.01.20 |



